Building structural models of biological molecules and study their interactions is at the core of my work. Here after are several examples of modelling studies I carried on. If interested, all my publications are listed in the Communications section.
Study of lipid - lipase interactions
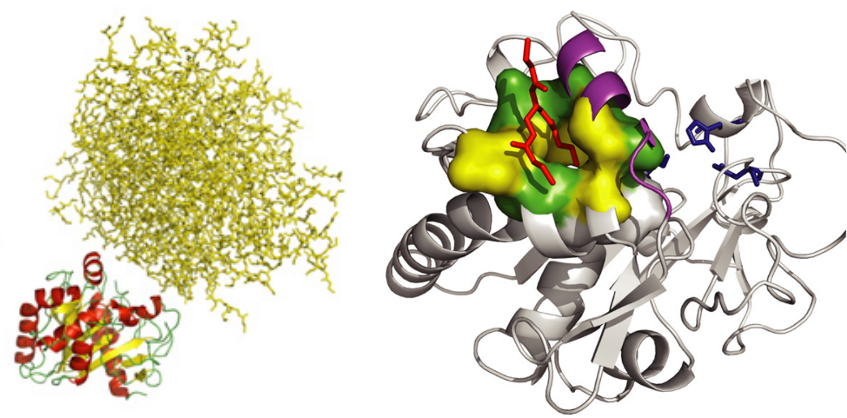
Thermomyces lanuginosa lipase has been widely studied in industrial and biotechnological research because of its potential for the transformation of triacylglycerol. We have studied the structure of this lipase by molecular dynamics in the presence of an aggregate of triglycerides. The simulations confirmed the high plasticity of the loop covering the active site (called LID) and showed that the lipid molecules also bind to a secondary pocket next to the LID. Together, these results strongly suggest that the LID plays a role in the anchoring of the protein to the aggregate. The simulations also revealed the existence of a polar channel that connects the active site to the solvent. This channel could regulate the water content of the active site. This study was published in the Biophysical Journal in 2009 (DOI: 10.1016/j.bpj.2009.03.040).
Study of Pseudodesmin peptide - peptide interactions
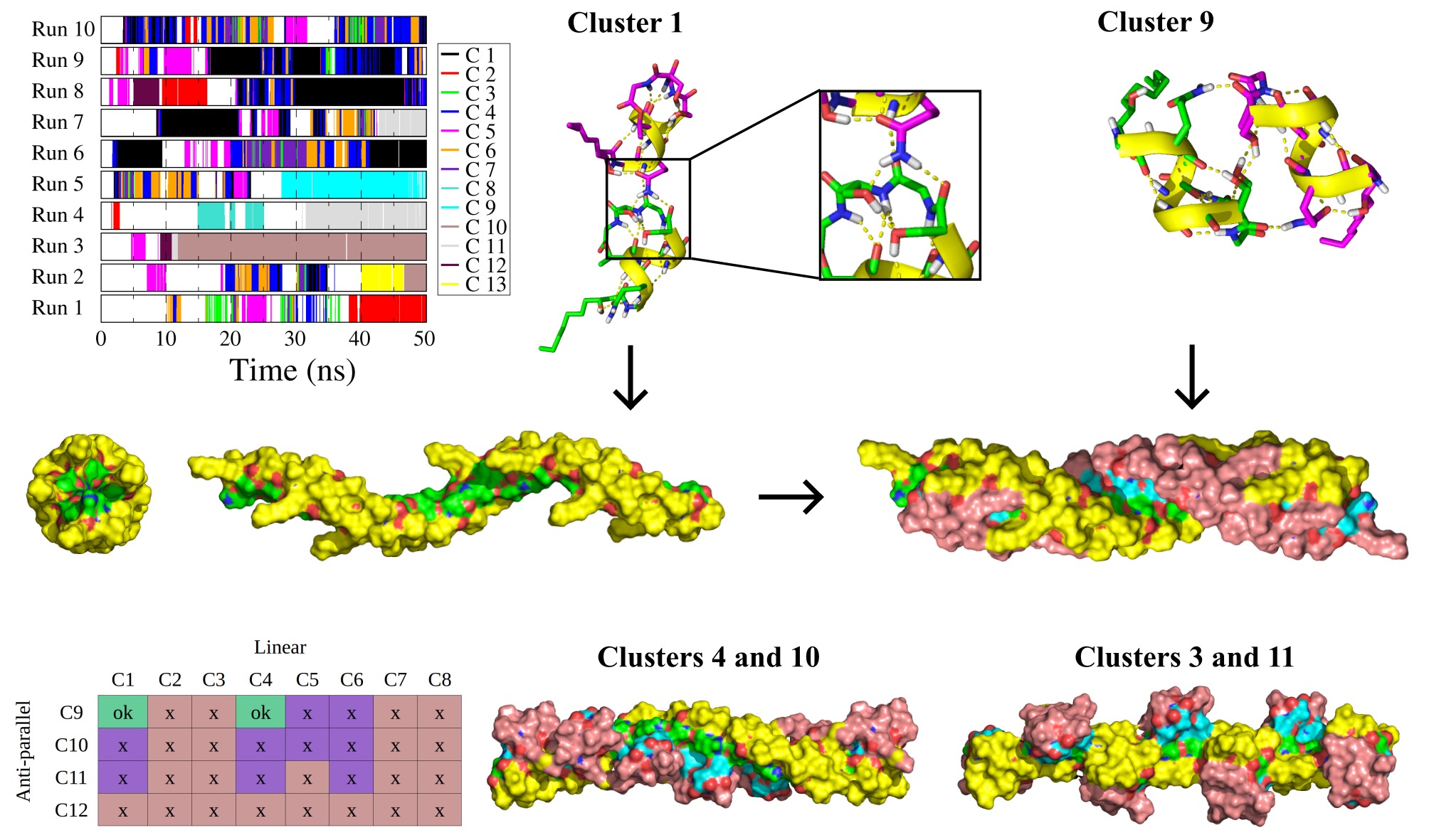
Pseudodesmine A is a cyclic lipodepsipeptide which exhibits moderate antibacterial activity and whose structure has been resolved by X-rays and NMR. This peptide, monomeric in acetonitrile, keeps the same structure in chloroform but assembles into a supramolecular complex. Comparison of the NMR data collected in the two solvents gave indications of the intermolecular contacts that occur but a structure of the agregate could not be defined. To study this assembly, dimerization simulations by molecular dynamics were carried out. Dimeric structures with linear, perpendicular and side by side configurations were observed. Those dimers were characterized by 2 to 4 hydrogen bonds. By combining linear and lateral dimers, it was possible to reconstruct filaments whose properties correspond to the values measured in NMR with respect to the width, the length, the orientation and the interaction of the peptides within the oligomers. This study was published in J Phys Chem B in 2019 (DOI: 10.1016/j.bpj.2009.03.040).
Research and study of peptides with the XGXPGXGXG motif of elastin
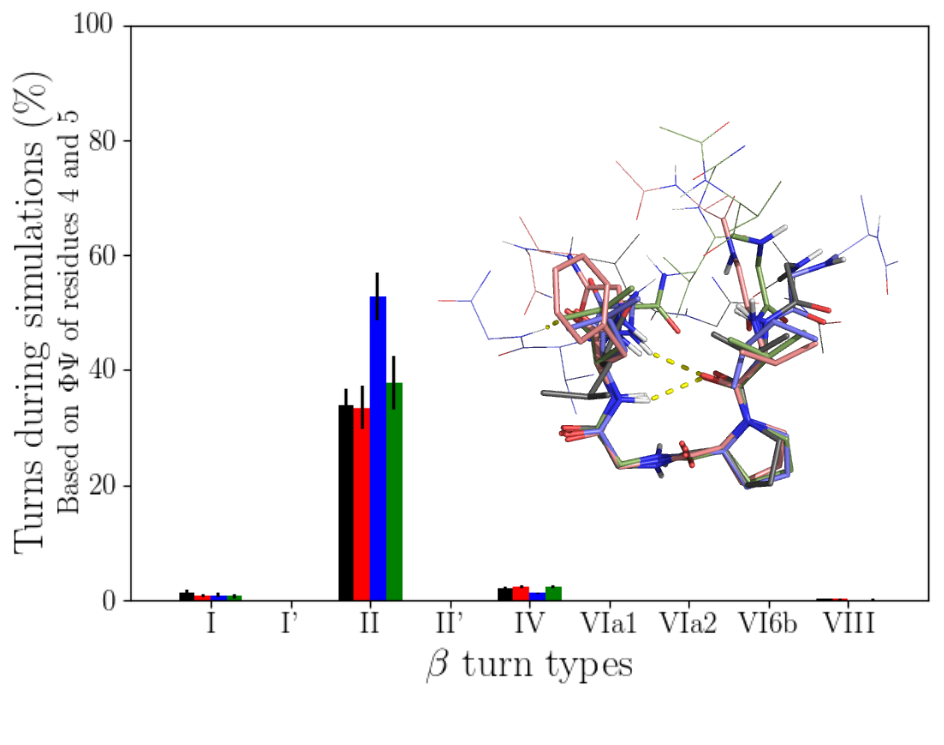
Peptides exhibiting this motif were searched for in the UniProtKB database. The results show that elastin peptides have particular characteristics. Indeed, in elastin, several motifs are often found in the same sequence and mostly in tandem. In addition, the variable residues of the motif are enriched in hydrophobic residues and some human elastin peptides are unique among the peptides found (i.e. AGVPGFGVG). The conformational characterization was then carried out by molecular dynamics on 4 human nonapeptides. The results show that the peptides are mainly in a “random coil” conformation and with a type II β turn. These β turns are formed at the level of the PG residues. Other types of β turns can also be formed in smaller amounts at other positions in the peptide sequence. There is good agreement between the spectcroscopic studies (CD and Raman) which were also carried out during this study and the simulations. CD spectra were notably calculated from MD simulations and compared to experimental spectra. This conformation could be relevant for the observed bioactivity of these peptides, in particular tumor progression, and their potential interaction with the ribosomal protein SA. This study has been published in the Biophysical Journal in 2020 (DOI: 10.1016/j.bpj.2020.04.019).
As membranes constitute the interface between living cells, organites and their environment, membrane interactions are of paramount importance in many biological processes as well as for biotechnological applications. Molecular dynamics gives the atomistic details of these processes and is very useful to study the evolution of bio-molecule and membrane structures following their interactions.
The SIV fusion peptide
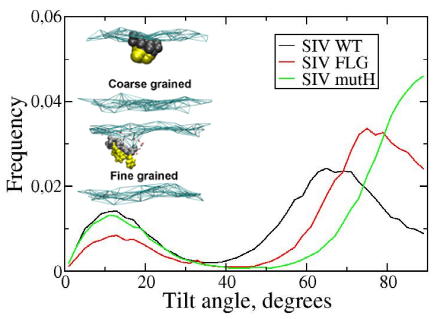
Fusion peptides are building blocks of several enveloped viruses that allow fusion between the host membrane and the viral membrane. It is generally suggested that these peptides can promote early fusion steps by inducing membrane curvature and that they occur as tilted α-helices in membranes. Although this property has been the subject of several experimental and in silico studies, extensive sampling of the peptide-membrane interaction has not yet been performed. In this study, we performed coarse-grained molecular dynamics simulations in which the lipid bilayer self-assembles around the peptide. The simulations indicate that the SIV fusion peptide can take on two different orientations in a model lipid bilayer, a major population which adopts a tilted interfacial orientation and a minor population which is perpendicular to the bilayer. The simulations also indicate that for the SIV mutant which does not induce fusion in vitro, the tilt is abolished. This study was published in Journal of Physical Chemistry B in 2012 (DOI: 10.1021/jp3027385).
The C-terminal domain of remorin
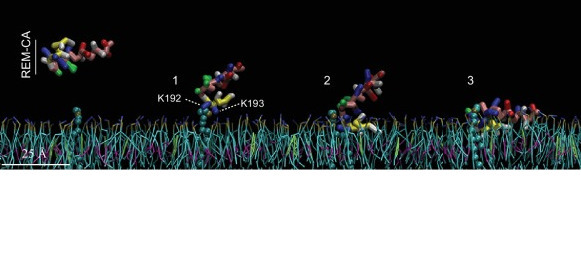
Remorin is a plant specific protein that is involved in viral resistance and recognized as a marker for lipid rafts in plants. Analysis of the sequence of this protein has identified a C-terminal membrane domain (RemCA) which has been shown to be essential and sufficient for the binding of the protein to the plasma membrane. This domain consists of an amphipathic α helix and an unstructured hydrophobic segment, the structures and behavior of which have been studied experimentally and by modeling, in particular in relation to the interaction with PIP4P. Amphipathy reducing mutations abolished the localization of remorin at the plasma membrane and its function in restricting the movement of potato virus X. This study resulted in two publications in Plant Physiology in 2012 (DOI: 10.1104/pp.112.200519) and Elife in 2017 (DOI: 10.7554/eLife.26404).
The C2 domains of MCTP
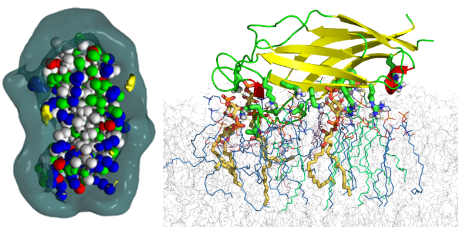
The detailed structure of plasmodesmas, which correspond to intercellular connections between cells of plants, is not yet fully understood. Within these structures, the transmembrane region of MCTPs could bind to the membrane of the endoplasmic reticulum while several C2 domains could interact with the plasma membrane. The objective of the work was to predict the membrane interaction capacities of these C2 domains. Sequence analysis was used to delineate the domains which were then modeled by homology modelling and then their membrane interaction was simulated. The results of these simulations allowed us to predict their modes of interaction and to underline the importance of electrostatic interactions with the polar heads of lipids. A specific interaction between the C2D domain and PI4.5P has been demonstrated. This study was published in EMBO Reports in 2019 (DOI: 10.15252/embr.201847182).