Writing scripts and informatic code is part of modelling. The following sections presents some of the developping work I have done.
MHP have been developped by Robert Brasseur in 1991 (PubMed ID: 1714906) and allows to represent hydrophobic properties of molecules. MHP decrease exponentially with distance and are computed from transfert energy associated to seven atoms types. MHP are presented as isopotential surfaces. Have a look to the paper for a more detailed description.
During my PhD at the laboratory of Robert Brasseur, I wrote a plugin for Pymol which calculates MHP from 3D files in PDB format. It has notably be used to follow the trajectory of CADY peptide (DOI: 10.1016/j.bbamem.2012.09.006). Laurence Lins also studied protein surfaces throuh MHP in 2003 (DOI: 10.1110/ps.0304803).
Source code: https://github.com/jmcrowet/mhp
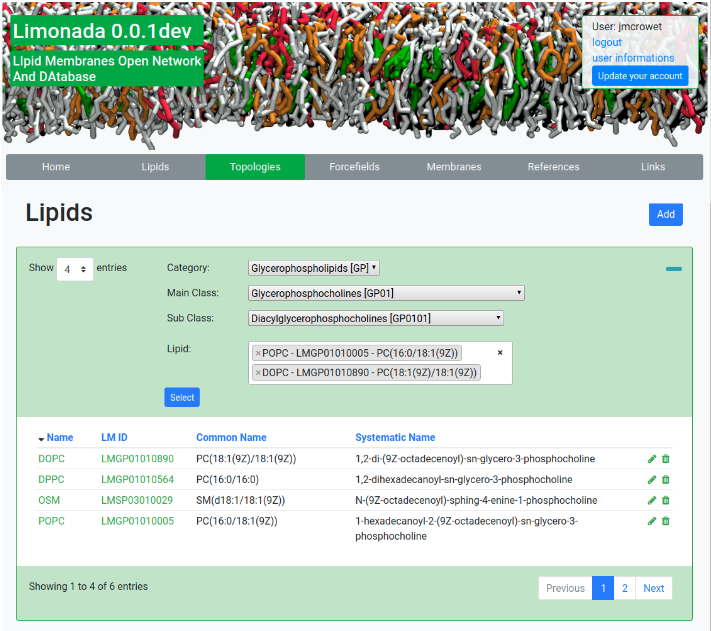
The size and complexity of lipid systems simulated by molecular dynamics is continuously growing. Knowing that there could be over 100,000 lipid species and that biological membranes can be composed by hundreds of different lipids whose proportion are organism and organelle specific, LIMONADA has been built as an open web database to provide model compositions representative of biological membranes, along with the description of lipids, topologies and force fields mandatory for their molecular dynamics simulations.
Website: https://limonada.univ-reims.fr/
Source code: https://github.com/LIMONADAMD